| Citation: | Chen Fei, Li Yuancheng, Qin Na, Wang Fengliang, Du Jiangbo, Wang Cheng, Du Fangzhi, Jiang Tao, Jiang Yue, Dai Juncheng, Hu Zhibin, Lu Cheng, Shen Hongbing. RNA-seq analysis identified hormone-related genes associated with prognosis of triple negative breast cancer[J]. The Journal of Biomedical Research, 2020, 34(2): 129-138. DOI: 10.7555/JBR.34.20190111 |
Breast cancer is the most diagnosed cancer and the leading cause of cancer-related death among females in the world, accounting for 25% of all cancer cases and 15% of all cancer deaths in women[1]. The incidence and mortality in China grew steadily in the last two decades[2], and the newly diagnosed breast cancer patients in China account for 12.2% of all newly diagnosed breast cancers worldwide, with 9.6% of deaths occurring in China[3]. Breast cancer is a heterogeneous disease with a variety of diversity in histologic, molecular and biological features[4], affecting the diagnosis and prognosis of breast cancer. Multiple molecular mechanisms involved in the course of it[5]. Triple negative breast cancer (TNBC) is a special subgroup characterized by lack of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression. As patients diagnosed with TNBC cannot respond to established anti-hormone and anti-HER2 therapies[6–7], chemotherapy remains the only standard therapeutic approach. Although some new chemical regimens and target agents have been developed, the efficacy of these regimens in the treatment of TNBC remains unclear. Additionally, since TNBC holds many aggressive features compared with other subtypes of breast cancer, such as susceptibility in younger women, shorter period to relapse and higher tendency to metastasize to viscera rather than to bone[7– 8], there is an urgent need to investigate potential biomarkers in the diagnosis and treatment of TNBC.
In addition to traditional clinicopathologic characteristics such as tumor size, the number of involved lymph nodes and fundamental biomarkers, microarray analysis has provided evidence that gene expression is important in the molecular classification and prognostic evaluation of breast cancer. Based on gene expression patterns, breast cancer can be classified into four main intrinsic subtypes including luminal-like, basal-like, HER2+ and normal breast[9–10]. And several gene expression signatures such as Mamma-Print[11] and Oncotype DX[12] have been developed to help predict the absolute benefit from a certain therapeutic regimen[13]. However, these signatures are valuable in patients with ER-positive breast cancer and less informative in ER-negative, high-proliferating breast tumors including TNBC[14]. Molecular mechanisms underlying carcinogenesis of TNBC still need to be further explored to discover better prognostic markers and new therapeutic targets. In recent years, RNA sequencing has emerged as a new technology for high throughput transcriptome analysis[15]. Many studies have identified that differentially expressed transcripts, spliced genes, and splice isoforms can be potential biomarkers in the classification of breast cancer subtypes, which will be beneficial for the diagnosis and treatment of breast cancer[16–17].
In this study, we performed a comprehensive differential analysis of TNBC samples by integrating RNA-seq data from both our cohort and The Cancer Genome Atlas (TCGA). We included both paired TNBC and adjacent non-cancerous tissues as well as other breast cancer subtypes, aiming to detect a group of differentially expressed genes (DEGs) that could universally represent the molecular features of TNBC and be helpful in searching for prognostic and predictive markers of TNBC.
Five paired breast tumors and matched adjacent normal tissues were obtained during surgical resection from the Maternity and Child Care Hospital of Nanjing Medical University. All tissues were snap-frozen. Informed consent was provided by all the subjects included in this study, which was approved by the Institutional Review Board of Nanjing Medical University. The tumor samples were microscopically confirmed to contain ≥70% invasive breast cancer cells by two independent pathologists, and the adjacent normal tissues contained no tumor cells. TNBC was confirmed according to the status of ER, PR, and HER2 based on immunohistochemical (IHC) classification. Briefly, tumors were considered negative for hormone receptors (ER and PR) if less than 1% of the tumor cells showed nuclear staining[18]. HER2 status was evaluated based on the criteria from the HER2 testing guideline[19]. The negative status of HER2 was defined as HER2 IHC 0 or 1+; samples with HER2 IHC 2+ should be subsequently subjected to in situ hybridization (ISH). Dako Envision immunohistochemistry staining system (Dako, Denmark) was employed for IHC testing. The primary antibodies were as follows: FLEX Monoclonal Rabbit Anti-Human Estrogen Receptor α (clone EP1, ready-to-use, Dako), FLEX Monoclonal Rabbit Anti-Human Progesterone Receptor (clone PgR636, ready-to-use, Dako), anti-HER2/neu (4B5) Rabbit Monoclonal Primary Antibody (VENTANA, USA). PathVysion HER2 DNA Probe Kit (Vysion, USA) was used for ISH assays. All the collected tissues were preserved in RNAlater solution (Ambion, USA) and stored at –80 °C prior to RNA extraction. The anamnestic and clinical-pathological characteristics of the 5 TNBC patients were shown in Supplementary Table 1 (available online). Total RNA was extracted from frozen breast cancer tissue samples using the RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. RNA pellets were resuspended in nuclease-free water, and the concentration was quantified using the RNA BR Assay kit for the Qubit 2.0 fluorimeter (Life Technologies, UK).
RNA quality was evaluated using the Agilent 2100 Bioanalyzer system (Agilent Technologies, USA). Only high-quality RNAs (RIN≥7.5) were selected for cDNA library construction subsequently. The cDNA libraries were prepared using the TruSeq RNA sample prep kit (Illumina, USA) and subsequently amplified in flowcells using Illumina cBot. Finally, standard Illumina RNA-seq protocol according to the manufacturer was conducted to generate the transcription profiles of the samples using the Illumina HiSeq 1500 (Illumina) for 2×101 cycles.
The FastQC version 0.11.8 was used to access the quality score distribution of the sequencing reads, and the low-quality reads (Phred score ≤20) were removed using Trimmomatic V0.32 prior to the analysis. The remaining qualified reads were aligned to the GENCODE Version 19 genome assembly using the TopHat (version 2.1.0) and BOWTIE (version 2.3.4.3), allowing two mismatches in the alignment for each read. Differential expression analysis of TNBC was performed with R 3.1.1 Bioconductor package DESeq2. And the batch effects were adjusted by Bioconductor package sva. URLs of the software mentioned above were presented in Supplementary Table 2 (available online).
We obtained the expression data and clinical information of breast cancers from TCGA Firehose Broad GDAC (http://gdac.broadinstitute.org/, version 2016-01-28 release). The breast cancer types, TNBC, ER+, and HER2+, were classified based on the IHC results included in the clinical data. Briefly, ER+ were defined as samples with ER positive, HER2 negative, and PR positive or negative, HER2+ samples were those with HER2 positive, ER and PR positive or negative, and TNBC samples were negative for all three receptors (ER/PR/HER2 all negative). In all, we included 595 ER+ breast cancer samples, 185 HER2+ breast cancer samples and 160 TNBC samples (including 14 samples with paired adjacent non-cancerous tissues available) in the following analysis. Clinical information on the cases obtained from the TCGA database was presented in Supplementary Table 3 (available online).
Differential expression analysis of TNBC was performed with R 3.1.1 Bioconductor package DESeq2. The following workflow was carried out for the comparison of TNBC tumor tissues versus non-tumor tissues or other two breast cancer types. We first removed genes with normalized read counts <5 in less than 20% samples. Raw read counts were normalized by DESeq2. The magnitude (log2 transformed fold change) and significance (P-value) of differential expression between groups were calculated, and genes with false discovery rate (FDR) adjusted P-values <0.05 were considered as significant. We obtained DEGs with the following criteria: FDR<0.05 and |log2 fold change|≥1. Genes significantly differentially expressed between TNBC tumor samples and both adjacent normal samples and other two breast cancer types were defined as TNBC-specific expressed genes.
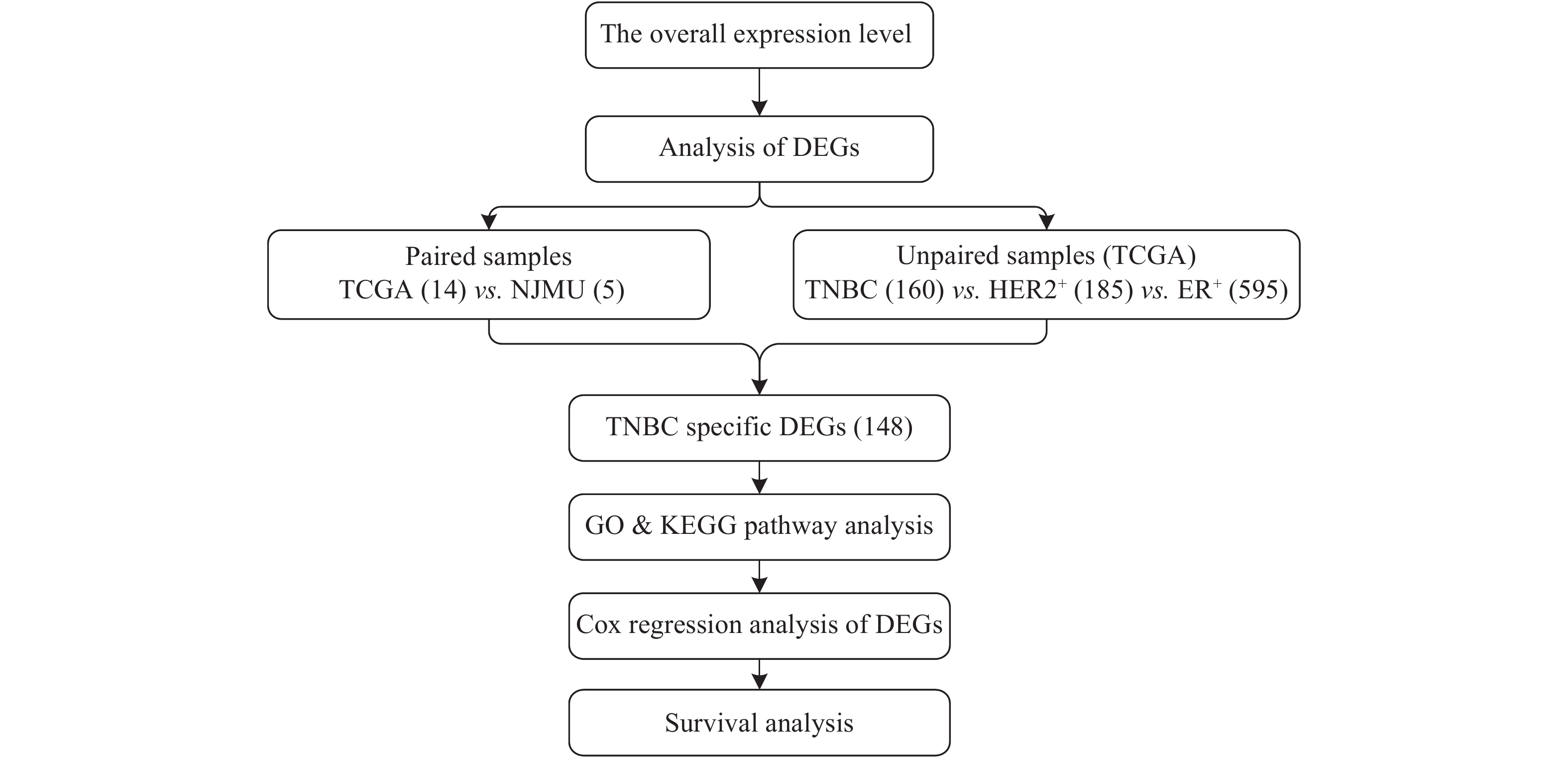
A pathway enrichment analysis was performed using the R 3.1.1 Bioconductor package clusterProfiler for the candidate TNBC-specific differential expressed genes. For multiple test correction, P-values were adjusted by the Benjamini-Hochberg false discovery rate (BH-FDR). The workflow was shown in Fig. 1.
Kaplan-Meier method and multivariate Cox proportional hazard regression analyses were performed to evaluate the association between expression of TNBC-specific expressed genes and breast cancer prognosis, with adjustment of age and clinical stage. Patients were divided into two groups: patients with gene expression above the median were defined as "high expression group", which represented a group of patients expressed relative high gene expression, and patients with gene expression below median level were defined as "low expression group". The crude or adjusted hazard ratio (HR) and their 95% confidence intervals (95% CI) were calculated. All tests were two-sided and all the significance level was set at FDR<0.05. We used the p. adjust function (based on BH-FDR) in R software to adjust P values. All the statistical analyses were carried out with R 3.1.1 software (http://www.cran.r-project.org/).
A total of 347.3 and 357.9 million reads were obtained from 10 independent libraries of paired breast cancer and adjacent non-cancerous tissues, respectively. Most reads reached Phred-like quality scores (Q-scores) at the Q30 level, indicating that the probability of an incorrect base call is 0.001%. The average coverage of sequencing depth reached approximately 53.45× of the human transcriptome. After alignment using TopHat and BOWTIE, 98.57% to 99.12% uniquely aligned reads of five paired tissues could be mapped to the human reference genome (Supplementary Table 4, available online).
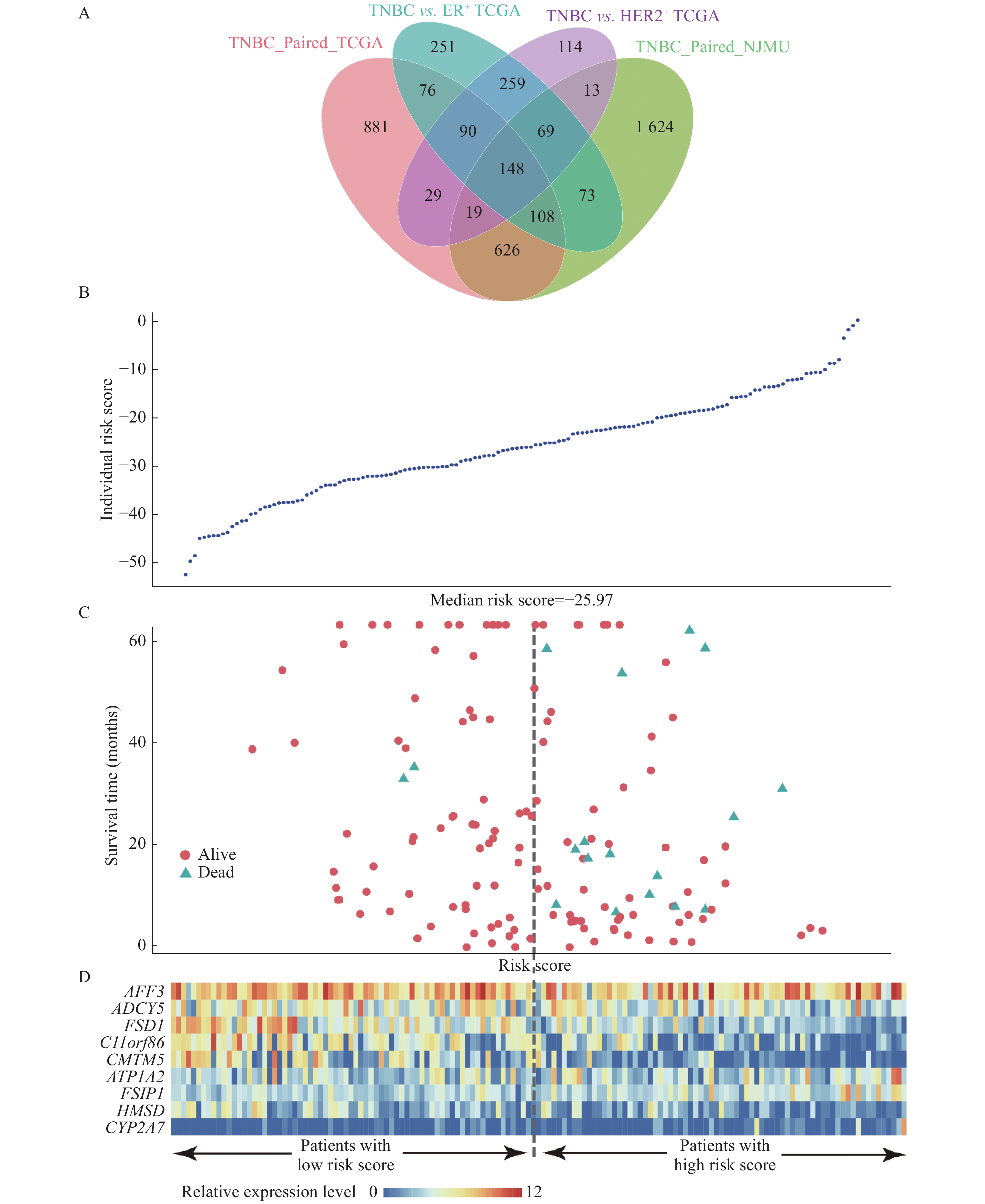
We first performed paired differential gene expression analysis, and 2 680 genes were found to be differentially expressed in 5 paired TNBC samples from our cohort, 1 977 genes were found in 14 paired TCGA samples and 894 genes were found in the same differential direction in the above two differentially expressed gene sets. Then, we further compared the transcriptome profiles between 160 TNBC samples and 595 ER+ and 185 HER2+ breast cancer samples respectively. We identified 741 and 1 074 DEGs between TNBC and the other two subtypes respectively. By integrating the tumor-adjacent differential expressed genes defined above, we identified a total of 148 TNBC-specific expressed genes [Fig. 2, Supplementary Fig. 1 and Supplementary Table 5 (available online)].
To evaluate the potential functions of the 148 TNBC-specific expressed genes defined in our study, we further performed enrichment analysis with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms. We identified that these genes were significantly enriched in 5 KEGG pathways and 33 GO terms. Interestingly, most of the enriched pathways and GO terms were related to the mammary gland morphogenesis and hormone levels related pathways, such as mammary gland duct morphogenesis, mammary gland epithelium development, steroid metabolic process and its like, suggesting that mammary gland morphogenesis might play a unique role in TNBC patients compared from other breast cancer types (Table 1 and 2).
| KEGG pathway | Count | Size | Adjusted P-value1 |
| Retinol metabolism | 5 | 64 | 5.70E-03 |
| Drug metabolism-cytochrome P450 | 5 | 73 | 5.70E-03 |
| Drug metabolism-other enzymes | 4 | 52 | 1.10E-02 |
| Caffeine metabolism | 2 | 7 | 1.56E-02 |
| PPAR signaling pathway | 4 | 70 | 2.03E-02 |
| Vascular smooth muscle contraction | 4 | 116 | 9.86E-02 |
| Nitrogen metabolism | 2 | 23 | 9.86E-02 |
| Ascorbate and aldarate metabolism | 2 | 26 | 9.86E-02 |
| Bile secretion | 3 | 71 | 9.86E-02 |
| Pentose and glucuronate interconversions | 2 | 32 | 1.31E-01 |
| 1adjusted by BH-FDR. | |||
| GO term | Count | GeneRatio | BackgroundRatio | Adjusted P value1 |
| Mammary gland duct morphogenesis | 5 | 5/125 | 31/17 381 | 5.22E-03 |
| Mammary gland epithelium Development | 6 | 6/125 | 66/17 381 | 7.87E-03 |
| Steroid metabolic process | 11 | 11/125 | 309/17 381 | 8.65E-03 |
| Mammary gland morphogenesis | 5 | 5/125 | 45/17 381 | 8.65E-03 |
| Lung epithelial cell differentiation | 4 | 4/125 | 24/17 381 | 9.61E-03 |
| Lung cell differentiation | 4 | 4/125 | 25/17 381 | 9.61E-03 |
| Prostate gland epithelium morphogenesis | 4 | 4/125 | 26/17 381 | 9.68E-03 |
| Regulation of hormone levels | 13 | 13/125 | 486/17 381 | 9.68E-03 |
| Lung secretory cell differentiation | 3 | 3/125 | 10/17 381 | 9.68E-03 |
| Prostate gland morphogenesis | 4 | 4/125 | 28/17 381 | 9.68E-03 |
| 1adjusted by BH-FDR. | ||||
Then, we further evaluate the prognostic effect of 148 TNBC-specific expressed genes in 145 TNBC cases from TCGA with survival information available. Kaplan-Meier method and multivariate Cox proportional hazard regression were performed. The base-line and follow-up information of the 145 patients was shown in Supplementary Table 6 and Supplementary Fig. 2 (available online). Interestingly, nine genes were found to be significantly associated with prognosis of TNBC patients, including FSIP1 (HR=4.61, P=6.92E−03), ADCY5 (HR=0.22, P=7.53E−03), FSD1 (HR=0.20, P=8.11E−03), HMSD (HR=0.24, P=1.13E−02), CMTM5 (HR=0.28, P=1.94E−02), AFF3 (HR=0.29, P=2.63E−02), CYP2A7 (HR=2.99, P=3.36E−02), ATP1A2 (HR=2.86, P=4.73E−02), and C11orf86 (HR=0.34, P=4.98E−02). Three of these nine genes were previously reported to be involved in the hormone-related pathways, including ADCY5, CYP2A7, and ATP1A2 [Table 3, Fig. 3, and Supplementary Fig. 3 (available online)].
| Gene Name | Ensembl Gene ID | TNBC | HER2+ | ER+ | |||||||||||
| HR | L95 | U95 | Padj1 | HR | L95 | U95 | Padj1 | HR | L95 | U95 | Padj1 | ||||
| FSIP1 | ENSG00000150667 | 4.61 | 1.52 | 13.99 | 6.92E-03 | 1.26 | 0.43 | 3.65 | 6.76E-01 | 0.38 | 0.16 | 0.89 | 2.58E-02 | ||
| ADCY5* | ENSG00000173175 | 0.22 | 0.07 | 0.67 | 7.53E-03 | 1.23 | 0.41 | 3.69 | 7.10E-01 | 0.75 | 0.34 | 1.62 | 4.60E-01 | ||
| FSD1 | ENSG00000105255 | 0.20 | 0.06 | 0.66 | 8.11E-03 | 0.49 | 0.16 | 1.52 | 2.19E-01 | 1.29 | 0.59 | 2.81 | 5.16E-01 | ||
| HMSD | ENSG00000221887 | 0.24 | 0.08 | 0.73 | 1.13E-02 | 0.18 | 0.02 | 1.44 | 1.07E-01 | 0.72 | 0.32 | 1.61 | 4.17E-01 | ||
| CMTM5 | ENSG00000166091 | 0.28 | 0.09 | 0.81 | 1.94E-02 | 1.03 | 0.34 | 3.06 | 9.64E-01 | 0.38 | 0.16 | 0.91 | 3.03E-02 | ||
| AFF3 | ENSG00000144218 | 0.29 | 0.10 | 0.86 | 2.63E-02 | 0.87 | 0.30 | 2.54 | 8.01E-01 | 0.90 | 0.43 | 1.88 | 7.71E-01 | ||
| CYP2A7* | ENSG00000198077 | 2.99 | 1.09 | 8.21 | 3.36E-02 | 0.75 | 0.25 | 2.23 | 6.00E-01 | 0.70 | 0.31 | 1.56 | 3.79E-01 | ||
| ATP1A2* | ENSG00000018625 | 2.86 | 1.01 | 8.05 | 4.73E-02 | 1.34 | 0.44 | 4.03 | 6.08E-01 | 0.56 | 0.25 | 1.24 | 1.55E-01 | ||
| C11orf86 | ENSG00000173237 | 0.34 | 0.12 | 0.99 | 4.98E-02 | 1.06 | 0.35 | 3.24 | 9.15E-01 | 1.20 | 0.55 | 2.62 | 6.51E-01 | ||
| DEGs: differentially expressed genes; HR: hazard ratio; L95: lower 95% confidence interval; U95: upper 95% confidence interval; 1Cox regression adjusted for age and AJCC pathologic tumor stage; *Hormone-related gene. | |||||||||||||||
TNBC accounts for approximately 15% of all invasive breast cancers and is recognized as an aggressive subgroup as it occurs primarily in young women and has a high potential to metastasize[7]. Currently, chemotherapy remains the mainstay of treatment for patients with TNBC for the absence of definite therapeutic targets, so it is of an urgent need to identify potential diagnostic and therapeutic markers such as immunotherapy[20]. Based on microarray and deep sequencing techniques, molecular alterations in TNBC have been widely explored including deficiency in homologous recombination, increased immunological infiltration, and expression of androgen receptors, leading to great progress in the clinical trials. However, the new drugs have been failed to be validated in phase 3 trials[21], leaving TNBC the most complex subset of breast carcinoma.
In the present study, we conducted a comprehensive differential expression analysis by comparing the expression profiles of TNBC samples with adjacent normal samples, ER+ breast cancers, and HER2+ breast cancers respectively. We identified a total of 148 TNBC-specific expressed genes, GO term enrichment analysis and KEGG pathway analysis found these genes were primarily enriched in mammary gland morphogenesis and hormone levels related pathways. Further survival analysis revealed that nine genes were significantly associated with the prognosis of TNBC patients.
GO term enrichment analysis suggested that most of the TNBC-specific expressed genes were enriched in steroid metabolic process and regulation of hormone levels. It has been well known that steroids induce both chronic and acute actions within cells, and mounting evidence shows that sex steroids are involved in cell growth and proliferation[22–23]. In some prior studies, steroid mediators (i.e. serum estrogens and androgens) were found to be associated with breast cancer risk[24]. Of note, estrogens might accelerate the development of breast cancer at many points along with the progression from early mutation to tumor metastasis. By increasing cell proliferation, estrogens may also decrease the capability of DNA damage repair[25]. Furthermore, estrogens might have genotoxic effects via their reactive metabolites[25]. Additionally, among TNBC patients, some investigators have reported that steroid Receptor Coactivator-3 performs as a therapeutic target due to its regulation of cellular signaling pathways that are vital for cancer proliferation and development, which can support our results as well[26]. Consequently, we can initially conclude that it is biologically plausible for us to observe the differential genes mainly involved in the steroid metabolic process in TNBC patients.
Among those genes, FSIP1, testicular and spermatogenesis-related antigen is expressed in abundance in various cancers, including breast cancer, lung cancer, etc. The level of FSIP1 is low or undetectable in most normal tissues except the testis[27]. In our study, we observed the prognostic value of FSIP1 in TNBC. High expression of FSIP1 was associated with a poorer outcome in TNBC, and reversely, a better outcome in ER+ patients. Consistent with our findings, several lines of evidence have previously indicated that FSIP1 expression in breast cancer correlated negatively with survival[28]. Indeed, FSIP1-mediated alterations in microtubule and dynein function may support the microtubule network and enhance mitotic robustness in cancer cells. Additionally, it is reported that FSIP1 can bind to HER2 directly in HER2+ breast cancer to promote cancer progression[27]. However, we didn't find a significant association between FSIP1 and prognosis in HER2+ breast cancer, which may due to a limited number of samples. As the correlation between ER status and FSIP1 was inconsistent among papers, larger sample studies and in vivo assays are needed to further validate[28–29].
CMTM5, a member of the CMTM protein family, is located at 14q11.2 and has been reported to act as a tumor-suppressor gene since it was specifically downregulated in numerous human cancers[30]. Previous studies also revealed that CMTM family proteins, including CMTM7, could inhibit cell growth and induce apoptosis[31]. Xu et al[30] reported that low expression of CMTM5 in hepatocellular carcinoma significantly correlated with poor overall survival. Interestingly, our study also suggested the prognostic value of CMTM5 and it might prolong the survival of patients with TNBC.
AFF3, a lymphoid-specific gene, encodes a 1227-amino acid protein that is presumed to play an important role in transcriptional regulation[32]. In our study, expression of AFF3 was observed to be associated with improved survival among patients with TNBC. Inconsistent with our results, AFF3 was showed to be related to worse overall survival in ER+ breast cancer treated with tamoxifen in previous studies[33]. No data to date is available regarding the role of AFF3 in TNBC. Hence, further investigation is required in terms of the biological mechanism and the prognostic value of AFF3 in TNBC.
ATP1A2 encodes the α2 subunit of the Na+/K+-ATPase (NKA) that is abundantly expressed in skeletal muscle and brain astrocytes. Some studies have demonstrated the anticancer effect of cardiac glycosides that directly inhibit NKA activity[34]. Down-regulation of ATP1A2 expression was previously reported in breast cancer[35]. In our study, we observed overexpression of ATP1A2 was associated with favorable outcome in TNBC. Given that the prognostic role of ATP1A2 in breast cancer remains unclear, more researches are required with respect to the effect of ATP1A2 on the occurrence and progression of breast cancer.
In addition, we observed that gene ADCY5, FSD1, HMSD, C11orf86 are down-regulated in TNBC with prolonging survival, whereas gene CYP2A7 is associated with poorer survival. To our knowledge, ADCY5 encodes adenylate cyclase 5, which catalyzes the generation of cAMP in type 2 diabetes[36]. FSD1 encodes a centrosome associated protein that is characterized by an N-terminal coiled-coil region downstream of B-box (BBC) domain, some studies found that the methylation of FSD1 may play a role in Epstein-Barr virus-associated gastric carcinomas[37]. CYP2A7 encodes a member of the cytochrome P450 superfamily, and cytochrome P450 metabolites are proved to play a critical role in carcinogenesis[38]. No data to date are available regarding the association between these genes and breast cancer. Thus, the results of our study are warranted to be further confirmed.
In conclusion, we identified a group of TNBC-specific expressed genes by comparing gene expression of TNBC tumors with that of paired normal tissues and non-TNBC tumors. Most of these genes were involved in the hormone-related pathways. Survival analysis further confirmed that nine steroid hormone-related genes were independent prognostic markers in TNBC. These findings emphasized the vital role of the hormone-related genes in TNBC tumorigenesis and may provide new therapeutic targets for TNBC.
We would like to thank all the people participated in this study. This work was supported by the Nanjing Medical Science and Technique Development Foundation (ZKX17041), the Natural Science Foundation of Jiangsu Province (BK20161120), and the Maternal and child health research project of Jiangsu Province (F201628), the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine) and Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015A067).
| [1] |
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012[J]. CA Cancer J Clin, 2015, 65(2): 87–108. doi: 10.3322/caac.21262
|
| [2] |
Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66(2): 115–132. doi: 10.3322/caac.21338
|
| [3] |
Fan L, Strasser-Weippl K, Li JJ, et al. Breast cancer in China[J]. Lancet Oncol, 2014, 15(7): E279–E289. doi: 10.1016/S1470-2045(13)70567-9
|
| [4] |
Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy?[J]. Nat Rev Clin Oncol, 2009, 6(12): 718–730. doi: 10.1038/nrclinonc.2009.166
|
| [5] |
Ni XJ, Zhao YC, Ma JJ, et al. Hypoxia-induced factor-1 alpha upregulates vascular endothelial growth factor C to promote lymphangiogenesis and angiogenesis in breast cancer patients[J]. J Biomed Res, 2013, 27(6): 478–485.
|
| [6] |
Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes[J]. Clin Cancer Res, 2007, 13(8): 2329–2334. doi: 10.1158/1078-0432.CCR-06-1109
|
| [7] |
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer[J]. N Engl J Med, 2010, 363(20): 1938–1948. doi: 10.1056/NEJMra1001389
|
| [8] |
Pogoda K, Niwińska A, Murawska M, et al. Analysis of pattern, time and risk factors influencing recurrence in triple-negative breast cancer patients[J]. Med Oncol, 2013, 30(1): 388. doi: 10.1007/s12032-012-0388-4
|
| [9] |
Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours[J]. Nature, 2000, 406(6797): 747–752. doi: 10.1038/35021093
|
| [10] |
Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications[J]. Proc Natl Acad Sci USA, 2001, 98(19): 10869–10874. doi: 10.1073/pnas.191367098
|
| [11] |
Cardoso F, van't Veer LJ, Bogaerts J, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer[J]. N Engl J Med, 2016, 375(8): 717–729. doi: 10.1056/NEJMoa1602253
|
| [12] |
Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer[J]. N Engl J Med, 2004, 351(27): 2817–2826. doi: 10.1056/NEJMoa041588
|
| [13] |
Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction[J]. Lancet, 2011, 378(9805): 1812–1823. doi: 10.1016/S0140-6736(11)61539-0
|
| [14] |
Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer[J]. Nat Rev Clin Oncol, 2017, 14(10): 595–610. doi: 10.1038/nrclinonc.2017.74
|
| [15] |
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics[J]. Nat Rev Genet, 2009, 10(1): 57–63. doi: 10.1038/nrg2484
|
| [16] |
Eswaran J, Cyanam D, Mudvari P, et al. Transcriptomic landscape of breast cancers through mRNA sequencing[J]. Sci Rep, 2012, 2: 264. doi: 10.1038/srep00264
|
| [17] |
Horvath A, Pakala SB, Mudvari P, et al. Novel insights into breast cancer genetic variance through RNA sequencing[J]. Sci Rep, 2013, 3: 2256. doi: 10.1038/srep02256
|
| [18] |
Hammond MEH, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer[J]. J Clin Oncol, 2010, 28(16): 2784–2795. doi: 10.1200/JCO.2009.25.6529
|
| [19] |
Wolff AC, Hammond MEH, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update[J]. J Clin Oncol, 2013, 31(31): 3997–4013. doi: 10.1200/JCO.2013.50.9984
|
| [20] |
Webb ES, Liu P, Baleeiro R, et al. Immune checkpoint inhibitors in cancer therapy[J]. J Biomed Res, 2018, 32(5): 317–326.
|
| [21] |
Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies[J]. Lancet, 2017, 389(10087): 2430–2442. doi: 10.1016/S0140-6736(16)32454-0
|
| [22] |
Louie MC, Sevigny MB. Steroid hormone receptors as prognostic markers in breast cancer[J]. Am J Cancer Res, 2017, 7(8): 1617–1636.
|
| [23] |
Bleach R, McIlroy M. The divergent function of androgen receptor in breast cancer; analysis of steroid mediators and tumor intracrinology[J]. Front Endocrinol (Lausanne), 2018, 9: 594. doi: 10.3389/fendo.2018.00594
|
| [24] |
The Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies[J]. J Natl Cancer Inst, 2002, 94(8): 606–616. doi: 10.1093/jnci/94.8.606
|
| [25] |
McNamara KM, Oguro S, Omata F, et al. The presence and impact of estrogen metabolism on the biology of triple-negative breast cancer[J]. Breast Cancer Res Treat, 2017, 161(2): 213–227. doi: 10.1007/s10549-016-4050-2
|
| [26] |
Song XZ, Zhang CW, Zhao MK, et al. Steroid Receptor Coactivator-3(SRC-3/AIB1) as a novel therapeutic target in triple negative breast cancer and its inhibition with a phospho-bufalin prodrug[J]. PLoS One, 2015, 10(10): e0140011. doi: 10.1371/journal.pone.0140011
|
| [27] |
Liu T, Zhang H, Sun L, et al. FSIP1 binds HER2 directly to regulate breast cancer growth and invasiveness[J]. Proc Natl Acad Sci USA, 2017, 114(29): 7683–7688. doi: 10.1073/pnas.1621486114
|
| [28] |
Chapman KB, Prendes MJ, Kidd JL, et al. Elevated expression of cancer/testis antigen FSIP1 in ER-positive breast tumors[J]. Biomark Med, 2013, 7(4): 601–611. doi: 10.2217/bmm.13.58
|
| [29] |
Zhang H, Luo MN, Jin ZN, et al. Expression and clinicopathological significance of FSIP1 in breast cancer[J]. Oncotarget, 2015, 6(12): 10658–10666.
|
| [30] |
Xu G, Dang CX. CMTM5 is downregulated and suppresses tumour growth in hepatocellular carcinoma through regulating PI3K-AKT signalling[J]. Cancer Cell Int, 2017, 17: 113. doi: 10.1186/s12935-017-0485-8
|
| [31] |
Li H, Li J, Su Y, et al. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth[J]. Oncogene, 2014, 33(24): 3109–3118. doi: 10.1038/onc.2013.282
|
| [32] |
Lefèvre L, Omeiri H, Drougat L, et al. Combined transcriptome studies identify AFF3 as a mediator of the oncogenic effects of β-catenin in adrenocortical carcinoma[J]. Oncogenesis, 2015, 4(7): e161. doi: 10.1038/oncsis.2015.20
|
| [33] |
Shi YW, Zhao Y, Zhang YJ, et al. AFF3 upregulation mediates tamoxifen resistance in breast cancers[J]. J Exp Clin Cancer Res, 2018, 37(1): 254. doi: 10.1186/s13046-018-0928-7
|
| [34] |
Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides[J]. Nat Rev Drug Discov, 2008, 7(11): 926–935. doi: 10.1038/nrd2682
|
| [35] |
Bogdanov A, Moiseenko FV, Dubina M. Abnormal expression of ATP1A1 and ATP1A2 in breast cancer[J]. F1000Res, 2017, 6: 10. doi: 10.12688/f1000research.10481.1
|
| [36] |
Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk[J]. Nat Genet, 2010, 42(2): 105–116. doi: 10.1038/ng.520
|
| [37] |
Okada T, Nakamura M, Nishikawa J, et al. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas[J]. Cancer Sci, 2013, 104(10): 1309–1314. doi: 10.1111/cas.12228
|
| [38] |
Modugno F, Knoll C, Kanbour-Shakir A, et al. A potential role for the estrogen-metabolizing cytochrome P450 enzymes in human breast carcinogenesis[J]. Breast Cancer Res Treat, 2003, 82(3): 191–197. doi: 10.1023/B:BREA.0000004376.21491.44
|
| [1] | Yingzhou Tu, Sen Wang, Haoran Wang, Peiyao Zhang, Mengyu Wang, Cunming Liu, Chun Yang, Riyue Jiang. The role of perioperative factors in the prognosis of cancer patients: A coin has two sides[J]. The Journal of Biomedical Research, 2025, 39(2): 117-127. DOI: 10.7555/JBR.38.20240164 |
| [2] | Jiaqi Tang, Lin Luo, Bakwatanisa Bosco, Ning Li, Bin Huang, Rongrong Wu, Zihan Lin, Ming Hong, Wenjie Liu, Lingxiang Wu, Wei Wu, Mengyan Zhu, Quanzhong Liu, Peng Xia, Miao Yu, Diru Yao, Sali Lv, Ruohan Zhang, Wentao Liu, Qianghu Wang, Kening Li. Identification of cell surface markers for acute myeloid leukemia prognosis based on multi-model analysis[J]. The Journal of Biomedical Research, 2024, 38(4): 397-412. DOI: 10.7555/JBR.38.20240065 |
| [3] | Wenqian Xia, Xiao Han, Lin Wang. E26 transformation-specific 1 is implicated in the inhibition of osteogenic differentiation induced by chronic high glucose by directly regulating Runx2 expression[J]. The Journal of Biomedical Research, 2022, 36(1): 39-47. DOI: 10.7555/JBR.35.20210123 |
| [4] | Xiao Shi, Xinxin Si, Ershao Zhang, Ruochen Zang, Nan Yang, He Cheng, Zhihong Zhang, Beijing Pan, Yujie Sun. Paclitaxel-induced stress granules increase LINE-1 mRNA stability to promote drug resistance in breast cancer cells[J]. The Journal of Biomedical Research, 2021, 35(6): 411-424. DOI: 10.7555/JBR.35.20210105 |
| [5] | Yang Ying, Chen Xiang, Ma Changyan. Insulin receptor is implicated in triple-negative breast cancer by decreasing cell mobility[J]. The Journal of Biomedical Research, 2021, 35(3): 189-196. DOI: 10.7555/JBR.34.20200082 |
| [6] | Liangyu Ge, Siyu Liu, Long Xie, Lei Sang, Changyan Ma, Hongwei Li. Differential mRNA expression profiling of oral squamous cell carcinoma by high-throughput RNA sequencing[J]. The Journal of Biomedical Research, 2015, 29(5): 397-404. DOI: 10.7555/JBR.29.20140088 |
| [7] | Yong Ji, Mingfeng Zheng, Shugao Ye, Jingyu Chen, Yijiang Chen. PTEN and Ki67 expression is associated with clinicopathologic features of non-small cell lung cancer[J]. The Journal of Biomedical Research, 2014, 28(6): 462-467. DOI: 10.7555/JBR.27.20130084 |
| [8] | Yun Pan, Chongqi Sun, Mingde Huang, Yao Liu, Fuzhen Qi, Li Liu, Juan Wen, Jibin Liu, Kaipeng Xie, Hongxia Ma, Zhibin Hu, Hongbing Shen. A genetic variant in pseudogene E2F3P1 contributes to prognosis of hepatocellular carcinoma[J]. The Journal of Biomedical Research, 2014, 28(3): 194-200. DOI: 10.7555/JBR.28.20140052 |
| [9] | Songyu Cao, Cheng Wang, Xinen Huang, Juncheng Dai, Lingmin Hu, Yao Liu, Jiaping Chen, Hongxia Ma, Guangfu Jin, Zhibin Hu, Lin Xu, Hongbing Shen. Prognostic assessment of apoptotic gene polymorphisms in non-small cell lung cancer in Chinese[J]. The Journal of Biomedical Research, 2013, 27(3): 231-238. DOI: 10.7555/JBR.27.20130014 |
| [10] | Wenze Sun, Liping Song, Ting Ai, Yingbing Zhang, Ying Gao, Jie Cui. Prognostic value of MET, cyclin D1 and MET gene copy number in non-small cell lung cancer[J]. The Journal of Biomedical Research, 2013, 27(3): 220-230. DOI: 10.7555/JBR.27.20130004 |
| 1. | Shi Z, Jiang T, Sun X, et al. HDAC10 inhibits non-small-cell lung cancer cell ferroptosis through the microRNA-223-5p-SLC7A11 axis. Toxicol Res (Camb), 2024, 13(5): tfae164. DOI:10.1093/toxres/tfae164 |
| 2. | Zhang W, Lee A, Tiwari AK, et al. Characterizing the Tumor Microenvironment and Its Prognostic Impact in Breast Cancer. Cells, 2024, 13(18): 1518. DOI:10.3390/cells13181518 |
| 3. | Supplitt S, Karpinski P, Sasiadek M, et al. The analysis of transcriptomic signature of TNBC-searching for the potential RNA-based predictive biomarkers to determine the chemotherapy sensitivity. J Appl Genet, 2024. DOI:10.1007/s13353-024-00876-x. Online ahead of print |
| 4. | Duan SL, Jiang Y, Li GQ, et al. Research insights into the chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM): their roles in various tumors. PeerJ, 2024, 12: e16757. DOI:10.7717/peerj.16757 |
| 5. | Yang H, Lachtara EM, Ran X, et al. The RNA m5C modification in R-loops as an off switch of Alt-NHEJ. Nat Commun, 2023, 14(1): 6114. DOI:10.1038/s41467-023-41790-w |
| 6. | Pei Y, Zhang Z, Tan S. Current Opinions on the Relationship Between CMTM Family and Hepatocellular Carcinoma. J Hepatocell Carcinoma, 2023, 10: 1411-1422. DOI:10.2147/JHC.S417202 |
| 7. | Ulaganathan K, Puranam K, Mukta S, et al. Expression profiling of luminal B breast tumor in Indian women. J Cancer Res Clin Oncol, 2023, 149(15): 13645-13664. DOI:10.1007/s00432-023-05195-y |
| 8. | Bellini D, Milan M, Bordin A, et al. A Focus on the Synergy of Radiomics and RNA Sequencing in Breast Cancer. Int J Mol Sci, 2023, 24(8): 7214. DOI:10.3390/ijms24087214 |
| 9. | Jayarathna DK, Rentería ME, Batra J, et al. Integrative competing endogenous RNA network analyses identify novel lncRNA and genes implicated in metastatic breast cancer. Sci Rep, 2023, 13(1): 2423. DOI:10.1038/s41598-023-29585-x |
| 10. | Molina-Ortiz D, Torres-Zárate C, Santes-Palacios R. Human Orphan Cytochromes P450: An Update. Curr Drug Metab, 2022, 23(12): 942-963. DOI:10.2174/1389200224666221209153032 |
| 11. | Barata IS, Gomes BC, Rodrigues AS, et al. The Complex Dynamic of Phase I Drug Metabolism in the Early Stages of Doxorubicin Resistance in Breast Cancer Cells. Genes (Basel), 2022, 13(11): 1977. DOI:10.3390/genes13111977 |
| 12. | Jayarathna DK, Rentería ME, Batra J, et al. A supervised machine learning approach identifies gene-regulating factor-mediated competing endogenous RNA networks in hormone-dependent cancers. J Cell Biochem, 2022, 123(8): 1394-1408. DOI:10.1002/jcb.30300 |
| 13. | Yan X, Dai J, Han Y, et al. FSIP1 Is Associated with Poor Prognosis and Can Be Used to Construct a Prognostic Model in Gastric Cancer. Dis Markers, 2022, 2022: 2478551. DOI:10.1155/2022/2478551 |
| 14. | Li Y, Yuan F, Lin Z, et al. Construction of endogenous RNA regulatory network for colorectal cancer based on bioinformatics. Zhong Nan Da Xue Xue Bao Yi Xue Ban, 2022, 47(4): 416-430. DOI:10.11817/j.issn.1672-7347.2022.210532 |
| 15. | Zeng Y, Zhang X, Li F, et al. AFF3 is a novel prognostic biomarker and a potential target for immunotherapy in gastric cancer. J Clin Lab Anal, 2022, 36(6): e24437. DOI:10.1002/jcla.24437 |
| 16. | Yan C, Liu Q, Jia R. Construction and Validation of a Prognostic Risk Model for Triple-Negative Breast Cancer Based on Autophagy-Related Genes. Front Oncol, 2022, 12: 829045. DOI:10.3389/fonc.2022.829045 |
| 17. | Cheng X, Murthy SRK, Zhuang T, et al. Canady Helios Cold Plasma Induces Breast Cancer Cell Death by Oxidation of Histone mRNA. Int J Mol Sci, 2021, 22(17): 9578. DOI:10.3390/ijms22179578 |
| 18. | Kashyap D, Garg VK, Sandberg EN, et al. Oncogenic and Tumor Suppressive Components of the Cell Cycle in Breast Cancer Progression and Prognosis. Pharmaceutics, 2021, 13(4): 569. DOI:10.3390/pharmaceutics13040569 |
| 19. | Yang Y, Chen X, Ma C. Insulin receptor is implicated in triple-negative breast cancer by decreasing cell mobility. J Biomed Res, 2020, 35(3): 189-196. DOI:10.7555/JBR.34.20200082 |
| KEGG pathway | Count | Size | Adjusted P-value1 |
| Retinol metabolism | 5 | 64 | 5.70E-03 |
| Drug metabolism-cytochrome P450 | 5 | 73 | 5.70E-03 |
| Drug metabolism-other enzymes | 4 | 52 | 1.10E-02 |
| Caffeine metabolism | 2 | 7 | 1.56E-02 |
| PPAR signaling pathway | 4 | 70 | 2.03E-02 |
| Vascular smooth muscle contraction | 4 | 116 | 9.86E-02 |
| Nitrogen metabolism | 2 | 23 | 9.86E-02 |
| Ascorbate and aldarate metabolism | 2 | 26 | 9.86E-02 |
| Bile secretion | 3 | 71 | 9.86E-02 |
| Pentose and glucuronate interconversions | 2 | 32 | 1.31E-01 |
| 1adjusted by BH-FDR. | |||
| GO term | Count | GeneRatio | BackgroundRatio | Adjusted P value1 |
| Mammary gland duct morphogenesis | 5 | 5/125 | 31/17 381 | 5.22E-03 |
| Mammary gland epithelium Development | 6 | 6/125 | 66/17 381 | 7.87E-03 |
| Steroid metabolic process | 11 | 11/125 | 309/17 381 | 8.65E-03 |
| Mammary gland morphogenesis | 5 | 5/125 | 45/17 381 | 8.65E-03 |
| Lung epithelial cell differentiation | 4 | 4/125 | 24/17 381 | 9.61E-03 |
| Lung cell differentiation | 4 | 4/125 | 25/17 381 | 9.61E-03 |
| Prostate gland epithelium morphogenesis | 4 | 4/125 | 26/17 381 | 9.68E-03 |
| Regulation of hormone levels | 13 | 13/125 | 486/17 381 | 9.68E-03 |
| Lung secretory cell differentiation | 3 | 3/125 | 10/17 381 | 9.68E-03 |
| Prostate gland morphogenesis | 4 | 4/125 | 28/17 381 | 9.68E-03 |
| 1adjusted by BH-FDR. | ||||
| Gene Name | Ensembl Gene ID | TNBC | HER2+ | ER+ | |||||||||||
| HR | L95 | U95 | Padj1 | HR | L95 | U95 | Padj1 | HR | L95 | U95 | Padj1 | ||||
| FSIP1 | ENSG00000150667 | 4.61 | 1.52 | 13.99 | 6.92E-03 | 1.26 | 0.43 | 3.65 | 6.76E-01 | 0.38 | 0.16 | 0.89 | 2.58E-02 | ||
| ADCY5* | ENSG00000173175 | 0.22 | 0.07 | 0.67 | 7.53E-03 | 1.23 | 0.41 | 3.69 | 7.10E-01 | 0.75 | 0.34 | 1.62 | 4.60E-01 | ||
| FSD1 | ENSG00000105255 | 0.20 | 0.06 | 0.66 | 8.11E-03 | 0.49 | 0.16 | 1.52 | 2.19E-01 | 1.29 | 0.59 | 2.81 | 5.16E-01 | ||
| HMSD | ENSG00000221887 | 0.24 | 0.08 | 0.73 | 1.13E-02 | 0.18 | 0.02 | 1.44 | 1.07E-01 | 0.72 | 0.32 | 1.61 | 4.17E-01 | ||
| CMTM5 | ENSG00000166091 | 0.28 | 0.09 | 0.81 | 1.94E-02 | 1.03 | 0.34 | 3.06 | 9.64E-01 | 0.38 | 0.16 | 0.91 | 3.03E-02 | ||
| AFF3 | ENSG00000144218 | 0.29 | 0.10 | 0.86 | 2.63E-02 | 0.87 | 0.30 | 2.54 | 8.01E-01 | 0.90 | 0.43 | 1.88 | 7.71E-01 | ||
| CYP2A7* | ENSG00000198077 | 2.99 | 1.09 | 8.21 | 3.36E-02 | 0.75 | 0.25 | 2.23 | 6.00E-01 | 0.70 | 0.31 | 1.56 | 3.79E-01 | ||
| ATP1A2* | ENSG00000018625 | 2.86 | 1.01 | 8.05 | 4.73E-02 | 1.34 | 0.44 | 4.03 | 6.08E-01 | 0.56 | 0.25 | 1.24 | 1.55E-01 | ||
| C11orf86 | ENSG00000173237 | 0.34 | 0.12 | 0.99 | 4.98E-02 | 1.06 | 0.35 | 3.24 | 9.15E-01 | 1.20 | 0.55 | 2.62 | 6.51E-01 | ||
| DEGs: differentially expressed genes; HR: hazard ratio; L95: lower 95% confidence interval; U95: upper 95% confidence interval; 1Cox regression adjusted for age and AJCC pathologic tumor stage; *Hormone-related gene. | |||||||||||||||